Variational Autoencoder (VAE)
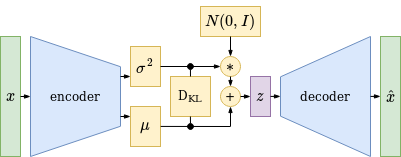 Variational Autoencoder Structure
Variational Autoencoder Structure
A Variational AutoEncoder is trained like a normal autoencoder to minimize the reconstruction loss, but in addition the latent space representation of the dataset is optimized to form a gaussian normal distribution with zero mean and unit variance. This can be done by using the Kullback-Leibler Divergence that calculates the distance between two probability distributions and allows to minimize this distance by gradient descent with respect to the encoder parameters. To get a robust latent space representation, latent space vectors are sampled using a gaussian distribution and the decoder is optimized to correctly map these sampled latent space vectors to the original target vectors. This adds a stochastic element to the learning process and prevents the VAE - together with the latent space regularisation - from overfitting. As a result - after training - the decoder can be used to generate new, better interpolated and more meaningful output data from unseen latent space vectors.
Deriving the Kullback-Leibler Divergence Loss
The Kullback-Leibler Divergence between the encoder output multivariate normal distribution
\[p(z \vert x) = \mathcal{N}(z \vert \mu_x, \Sigma_x) = \mathcal{N}(\mu_0, \Sigma_0)\]and the desired zero mean unit variance latent space normal distribution
\[p(z) = \mathcal{N}(0, \pmb{I}) = \mathcal{N}(\mu_1, \Sigma_1)\]is defined as
\[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \left( tr(\Sigma_1^{-1}\Sigma_0) + ln \left( \frac{\vert\Sigma_1\vert}{\vert\Sigma_0\vert} \right) - n + (\mu_1 - \mu_0)^T\Sigma_1^{-1}(\mu_1 - \mu_0) \right)\]with \(\mu_1 = 0\) and \(\Sigma_1 = \pmb{I}\) we get
\[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \left( tr(\pmb{I}^{-1}\Sigma_0) + ln \left( \frac{\vert\pmb{I}\vert}{\vert\Sigma_0\vert} \right) - n + (0 - \mu_0)^T\pmb{I}^{-1}(0 - \mu_0) \right)\] \[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \left( tr(\Sigma_0) - ln( \vert\Sigma_0\vert ) - n + \mu_0^T\mu_0 \right)\]with i as the n-dimensional latent space feature index
\[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \left( \sum_i \sigma_i^2 - ln( \prod_i \sigma_i^2 ) - n + \sum_i \mu_i^2 \right)\]we get the loss function based on the encoder output variance and mean vectors
\[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \left( \sum_i \sigma_i^2 - \sum_i ln( \sigma_i^2 ) - n + \sum_i \mu_i^2 \right)\] \[D_{KL}(\mathcal{N}_0 \vert\vert \mathcal{N}_1) = \frac{1}{2} \sum_i \left( \sigma_i^2 - ln( \sigma_i^2 ) - 1 + \mu_i^2 \right)\]The derivative of the loss function with respect to \(\sigma^2\) gives us the variance gradient
\[\frac{d D_{KL}}{d \sigma_i^2} = \frac{1}{2} \left( 1 - \frac{1}{\sigma_i^2} \right) = \frac{1}{2} - \frac{1}{2 \sigma_i^2}\]and the derivative of the loss function with respect to \(\mu\) gives us the mean gradient
\[\frac{d D_{KL}}{d \mu_i} = \frac{1}{2} \left( 2 \mu_i \right) = \mu_i\]Nomenclature
- x and z : encoder input vector and latent space vector.
- p : probability distribution.
- \(\mathcal{N}\) : a gaussian normal distribution, represented by an expected value vector and a covariance matrix.
- \(\pmb{I}\) : identity matrix.
- \(\Sigma\) : the covariance matrix of a probability distribution. In case of the VAE, the latent space covariance matrix is defined as a diagonal matrix and can therefore be represented by its diagonal vector. The non-diagonal elements are supposed to be 0 which implies that the latent space features are uncorrelated / independent of each other.
- \(\sigma^2\) : a variance vector = vector of squared standard deviation values.
- \(\mu\) : a mean vector.
- \(tr(A)\) : the trace of the matrix that equals the sum over all diagonal elements of the matrix.
- \(\vert A \vert\) : the determinant of the matrix that - in case of a diagonal matrix - equals the product over all diagonal elements of the matrix.
- \(ln\) : the natural log is a good choice when calculating the Kullback-Leibler Divergence of multivariate normal distributions.
To get meaningful variance values - which are always positive - the activation function which generates \(\sigma^2\) has to softly map negative values from the encoder network to positive values. A good choice is the Softplus activation function. This also prevents negative values to be fed into the log function which is part of the Kullback-Leibler Divergence loss. The mean vector \(\mu\) can simply be generated using a Linear activation function.
Variational Autoencoder Implementation
import numpy_neural_network as npnn
import npnn_datasets
from numpy_neural_network import Sequential
encoder_model = npnn.Sequential()
encoder_model.layers = [
npnn.Conv2D(shape_in=(3, 3, 1), shape_out=(2, 2, 6), kernel_size=2, stride=1),
npnn.Tanh(2 * 2 * 6),
npnn.Conv2D(shape_in=(2, 2, 6), shape_out=(1, 1, 2), kernel_size=2, stride=1),
npnn.Tanh(1 * 1 * 2),
npnn.Latent(shape_in=(1, 1, 2))
]
decoder_steps_per_encoder_step = 25
class DecoderSequential(Sequential):
# override of Sequential step method
def step(self, x=None, t=None):
global decoder_steps_per_encoder_step
g_dec = np.zeros(x.shape)
for _ in np.arange(decoder_steps_per_encoder_step):
x_dec = x.copy()
t_dec = t.copy()
x_dec = self.forward(x_dec)
g, y_dec = self.chain.step(x=x_dec, t=t_dec)
g_dec += self.backward(g)
g_dec = g_dec / decoder_steps_per_encoder_step
self.loss = self.chain.loss / decoder_steps_per_encoder_step
self.accuracy = self.chain.accuracy / decoder_steps_per_encoder_step
return g_dec, y_dec
decoder_model = DecoderSequential()
decoder_model.layers = [
npnn.Sample(shape_out=(1, 1, 2)),
npnn.UpConv2D(shape_in=(1, 1, 2), shape_out=(2, 2, 6), kernel_size=2, stride=1),
npnn.Tanh(2 * 2 * 6),
npnn.UpConv2D(shape_in=(2, 2, 6), shape_out=(3, 3, 1), kernel_size=2, stride=1),
npnn.Tanh(3 * 3 * 1),
npnn.Dense(shape_in=(3, 3, 1), shape_out=(3, 3, 1)),
npnn.Linear(3 * 3 * 1)
]
kl_loss_layer = npnn.loss_layer.KullbackLeiblerLoss(shape_in=(1, 1, 4))
loss_layer = npnn.loss_layer.RMSLoss(shape_in=(3, 3, 1))
optimizer = npnn.optimizer.Adam(alpha=5e-3)
dataset = npnn_datasets.FourSmallImages()
encoder_model.chain = kl_loss_layer
kl_loss_layer.chain = decoder_model
decoder_model.chain = loss_layer
optimizer.norm = dataset.norm
optimizer.model = encoder_model
Variational Autoencoder with Latent Space Regularisation using KL-Divergence
Variational Autoencoder References
 Ava Soleimany - Deep Generative Modeling, MIT 6.S191 (2020)
Ava Soleimany - Deep Generative Modeling, MIT 6.S191 (2020)
 Serena Yeung - Generative Models, Stanford University cs231n (2017)
Serena Yeung - Generative Models, Stanford University cs231n (2017)